-
Notifications
You must be signed in to change notification settings - Fork 12
Commit
This commit does not belong to any branch on this repository, and may belong to a fork outside of the repository.
Merge pull request #86 from gaelforget/v0p3p4d
V0p3p4d
- Loading branch information
Showing
29 changed files
with
10,101 additions
and
857 deletions.
There are no files selected for viewing
This file contains bidirectional Unicode text that may be interpreted or compiled differently than what appears below. To review, open the file in an editor that reveals hidden Unicode characters.
Learn more about bidirectional Unicode characters
This file contains bidirectional Unicode text that may be interpreted or compiled differently than what appears below. To review, open the file in an editor that reveals hidden Unicode characters.
Learn more about bidirectional Unicode characters
| Original file line number | Diff line number | Diff line change |
|---|---|---|
| @@ -1,18 +1,18 @@ | ||
| name = "IndividualDisplacements" | ||
| uuid = "b92f0c32-5b7e-11e9-1d7b-238b2da8b0e6" | ||
| authors = ["gaelforget <[email protected]>"] | ||
| version = "0.3.3" | ||
| version = "0.3.4" | ||
|
|
||
| [deps] | ||
| CFTime = "179af706-886a-5703-950a-314cd64e0468" | ||
| CSV = "336ed68f-0bac-5ca0-87d4-7b16caf5d00b" | ||
| CyclicArrays = "6023044f-b3ef-4c22-8399-3ec90a0c9b75" | ||
| DataFrames = "a93c6f00-e57d-5684-b7b6-d8193f3e46c0" | ||
| Dates = "ade2ca70-3891-5945-98fb-dc099432e06a" | ||
| LazyArtifacts = "4af54fe1-eca0-43a8-85a7-787d91b784e3" | ||
| MeshArrays = "cb8c808f-1acf-59a3-9d2b-6e38d009f683" | ||
| NetCDF = "30363a11-5582-574a-97bb-aa9a979735b9" | ||
| OrdinaryDiffEq = "1dea7af3-3e70-54e6-95c3-0bf5283fa5ed" | ||
| Pkg = "44cfe95a-1eb2-52ea-b672-e2afdf69b78f" | ||
| Random = "9a3f8284-a2c9-5f02-9a11-845980a1fd5c" | ||
| UnPack = "3a884ed6-31ef-47d7-9d2a-63182c4928ed" | ||
|
|
||
|
|
||
This file contains bidirectional Unicode text that may be interpreted or compiled differently than what appears below. To review, open the file in an editor that reveals hidden Unicode characters.
Learn more about bidirectional Unicode characters
| Original file line number | Diff line number | Diff line change |
|---|---|---|
| @@ -1,16 +1,19 @@ | ||
| [deps] | ||
| CSV = "336ed68f-0bac-5ca0-87d4-7b16caf5d00b" | ||
| CairoMakie = "13f3f980-e62b-5c42-98c6-ff1f3baf88f0" | ||
| ColorSchemes = "35d6a980-a343-548e-a6ea-1d62b119f2f4" | ||
| CyclicArrays = "6023044f-b3ef-4c22-8399-3ec90a0c9b75" | ||
| DataFrames = "a93c6f00-e57d-5684-b7b6-d8193f3e46c0" | ||
| Documenter = "e30172f5-a6a5-5a46-863b-614d45cd2de4" | ||
| Literate = "98b081ad-f1c9-55d3-8b20-4c87d4299306" | ||
| LiveServer = "16fef848-5104-11e9-1b77-fb7a48bbb589" | ||
| MITgcmTools = "62725fbc-3a66-4df3-9000-e33e85b3a198" | ||
| MeshArrays = "cb8c808f-1acf-59a3-9d2b-6e38d009f683" | ||
| NetCDF = "30363a11-5582-574a-97bb-aa9a979735b9" | ||
| OceanStateEstimation = "891f6deb-a4f5-4bc5-a2e3-1e8f649cdd2c" | ||
| OrdinaryDiffEq = "1dea7af3-3e70-54e6-95c3-0bf5283fa5ed" | ||
| Plots = "91a5bcdd-55d7-5caf-9e0b-520d859cae80" | ||
| PlutoSliderServer = "2fc8631c-6f24-4c5b-bca7-cbb509c42db4" | ||
| Random = "9a3f8284-a2c9-5f02-9a11-845980a1fd5c" | ||
| Statistics = "10745b16-79ce-11e8-11f9-7d13ad32a3b2" | ||
| StatsBase = "2913bbd2-ae8a-5f71-8c99-4fb6c76f3a91" |
This file contains bidirectional Unicode text that may be interpreted or compiled differently than what appears below. To review, open the file in an editor that reveals hidden Unicode characters.
Learn more about bidirectional Unicode characters
This file contains bidirectional Unicode text that may be interpreted or compiled differently than what appears below. To review, open the file in an editor that reveals hidden Unicode characters.
Learn more about bidirectional Unicode characters
| Original file line number | Diff line number | Diff line change |
|---|---|---|
| @@ -1,81 +1,56 @@ | ||
| ## Summary | ||
|
|
||
| The four examples outlined below form a tutorial of sort, and thus complement the rest of the package documentation. The corresponing data wrangling examples in `flow_fields.jl` define grids and ingest velocity fields. They hopefully provide a useful jumping off point in order to configure `IndividualDisplacements.jl` for new problems. | ||
| The four examples outlined below form a tutorial of sorts, and thus complement the rest of the package documentation. They rely on `flow_fields.jl` to define grids and ingest velocity fields. The main goal is to provide a useful jumping off point in order to configure `IndividualDisplacements.jl` for new problems. | ||
|
|
||
| Output is in [DataFrames](https://juliadata.github.io/DataFrames.jl/latest/) tabular format which comes with powerful and convenient analysis methods. Plotting results in space and time can be done as in `recipes_plots.jl`, `recipes_makie.jl`, and `recipes_pyplot.jl` -- see the examples. | ||
|
|
||
| ## Particle Set Example | ||
| To run an example, the recommended method is to copy the corresponding `notebook (code)` link, paste into the [Pluto.jl](https://github.com/fonsp/Pluto.jl/wiki/🔎-Basic-Commands-in-Pluto) prompt, and click `open`. | ||
|
|
||
| Simulation of an ensemble of displacements (and trajectories) in a simple 2D configuration. | ||
| ## Simple Two-Dimensional Flow | ||
|
|
||
| [notebook (html)](random_flow_field.html) ➭ [notebook (code)](https://github.com/JuliaClimate/IndividualDisplacements.jl/blob/master/examples/basics/random_flow_field.jl) | ||
|
|
||
| Simulate an ensemble of displacements (and trajectories) in a simple 2D configuration. | ||
|
|
||
| The `convert_to_FlowFields` convenience function defines a simplified gridded domain that matches the velocity array size, adds a time range, and returns a `FlowFields` | ||
| data structure `𝐹`. | ||
| All that is left to do at this stage is to define initial conditions, put them together with `𝐹` within the `Individuals` data structure `𝐼`, and call `∫!(𝐼)`. | ||
|
|
||
| The prototype for this example is based on a randomly generated flow field in a doubly periodic gridded domain. | ||
| Exercises include the non-periodic domain case, statistics made easy via `DataFrames.jl`, and replacing the flow field with your own. | ||
|
|
||
|  | ||
| ## Simple Three-Dimensional Flow | ||
|
|
||
| ## Single Particle Example | ||
| [notebook (html)](solid_body_rotation.html) ➭ [notebook (code)](https://github.com/JuliaClimate/IndividualDisplacements.jl/blob/master/examples/basics/solid_body_rotation.jl) | ||
|
|
||
| Setup a three-dimensional flow field `u,v,w`, initialize a single particle / individual position `📌`, and wrap everything up within an `Individuals` data structure `𝐼`. | ||
| Set up a three-dimensional flow field `u,v,w`, initialize a single particle at position `📌`, and wrap everything up within an `Individuals` data structure `𝐼`. | ||
|
|
||
| `𝐼` is displaced by integrating the individual velocity, [moving along through space](https://en.wikipedia.org/wiki/Lagrangian_and_Eulerian_specification_of_the_flow_field), over time `𝑇`. This is the main computation done in this package -- interpolating `u,v,w` to individual positions `𝐼.📌` on the fly, using `𝐼.🚄`, and integrating through time, using `𝐼.∫`. | ||
|
|
||
| The flow field consists of [rigid body rotation](https://en.wikipedia.org/wiki/Rigid_body), plus a convergent term, plus a sinking term in the vertical direction. This flow field generates a downward, converging spiral -- a idealized version of a relevant case in the Ocean. | ||
|
|
||
|  | ||
|
|
||
| ## Global Ocean Circulation | ||
|
|
||
| [notebook (html)](global_ocean_circulation.html) ➭ [notebook (code)](https://github.com/JuliaClimate/IndividualDisplacements.jl/blob/master/examples/worldwide/global_ocean_circulation.jl) | ||
|
|
||
| A simulation of floating particles over the Global Ocean which illustrates (1) using time variable velocity fields, (2) global connections, (3) particle re-seeding, and (4) output statistics. | ||
|
|
||
| The flow field is based on a data-constrained ocean model solution. The problem is configured in a way to mimic, albeit very crudely, the near-surface tranport of plastics or planktons. | ||
|
|
||
| [](https://youtu.be/W5DNqJG9jt0) | ||
|
|
||
| ## Three Dimensional Pathways | ||
|
|
||
| [notebook (html)](three_dimensional_ocean.html) ➭ [notebook (code)](https://github.com/JuliaClimate/IndividualDisplacements.jl/blob/master/examples/worldwide/three_dimensional_ocean.jl) | ||
|
|
||
| A simulation of particles that follow the three-dimensional ocean circulation. This example illustrates (1) the 3D case in a relatistic configuration, (2) tracking the advent or origin of a water patch, and (3) multifacted visualizations in 3D. | ||
|
|
||
| The flow field is based on a data-constrained, realistic, ocean model. The problem configuration mimics, albeit very approximately, ocean tracers / coumpounds transported by water masses . | ||
|
|
||
| [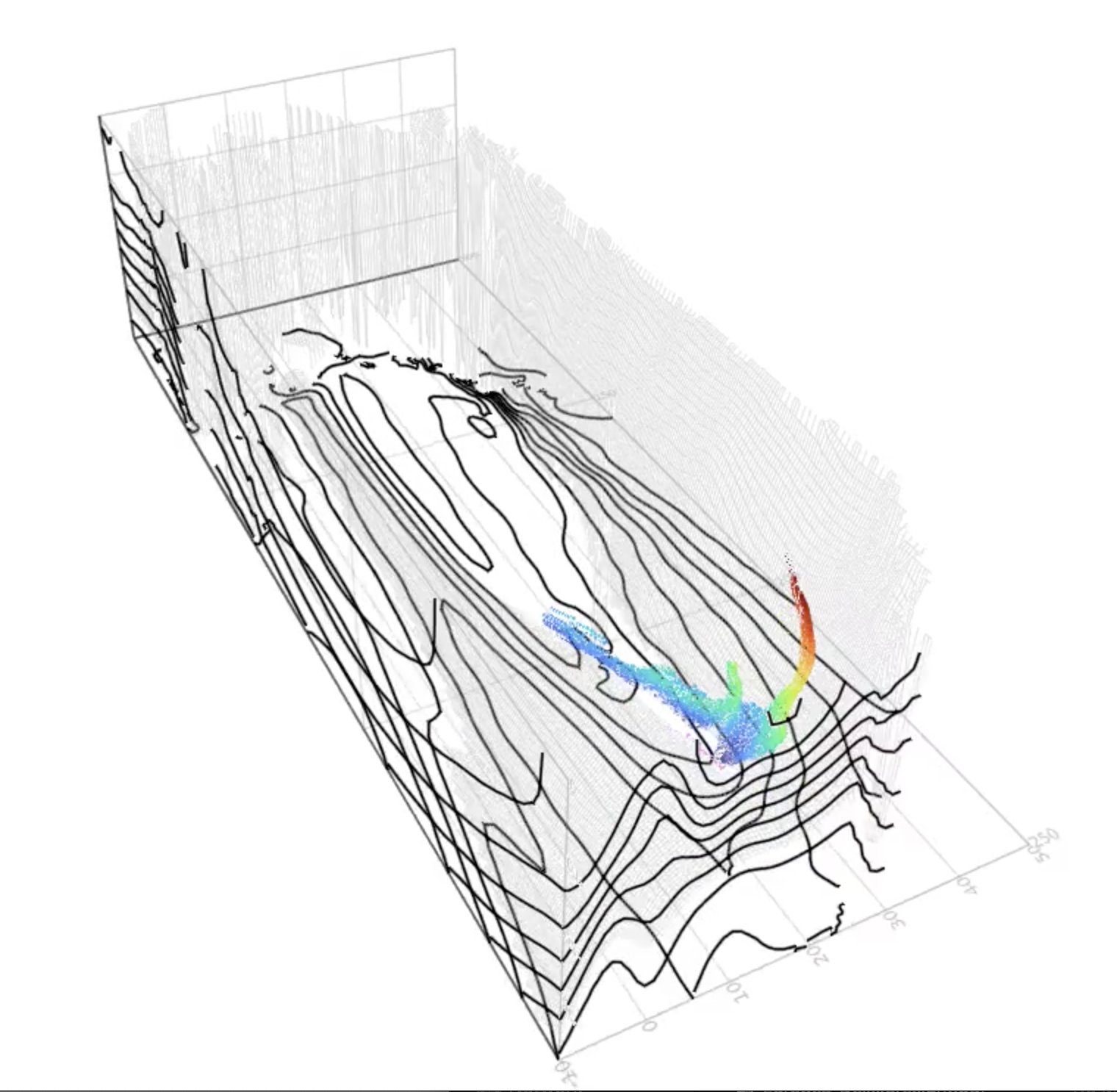](https://youtu.be/twAAE_WUs_g) | ||
|
|
||
| ## Running The Examples | ||
|
|
||
| Running the examples requires `julia` and its relevant packages. Inputs get downloaded as needed upon running the examples. The following commands ought to run on either your local computer or e.g [here in the cloud](https://mybinder.org/v2/gh/JuliaClimate/IndividualDisplacements.jl/master). | ||
|
|
||
| Maybe the simplest option is to generate jupyter notebooks for all examples as part of the documentation build (see below). The generated notebooks should be easy to re-run afterwards. To generate and run the notebooks, start `julia` in the terminal or a jupyter notebook at the `IndividualDisplacements.jl/` folder. Then try: | ||
| ## Additional Examples | ||
|
|
||
| ``` | ||
| using Pkg | ||
| Pkg.activate("docs/") | ||
| Pkg.instantiate() | ||
| Pkg.add(path="./") | ||
| include("docs/make.jl") | ||
| ``` | ||
|
|
||
| Or alternatively, to generate notebooks with more detailed control: | ||
| - Interactive UI (Pluto.jl) : [notebook (html)](interactive_UI.html) ➭ [notebook (code)](https://github.com/JuliaClimate/IndividualDisplacements.jl/blob/master/examples/worldwide/interactive_UI.jl) | ||
|
|
||
| ``` | ||
| using Literate | ||
| set1 = ["solid_body_rotation","random_flow_field","detailed_look","particle_cloud"] | ||
| [Literate.notebook("examples/basics/"*i*".jl", ".", execute = true, documenter = false) for i in set1] | ||
| set2 = ["global_ocean_circulation","three_dimensional_ocean"] | ||
| [Literate.notebook("examples/worldwide/"*i*".jl", ".", execute = true, documenter = false) for i in set2] | ||
| ``` | ||
| - Particle cloud (MITgcm) : [notebook (html)](../particle_cloud/index.html) ➭ [notebook (code)](https://github.com/JuliaClimate/IndividualDisplacements.jl/blob/master/examples/basics/particle_cloud.jl) | ||
|
|
||
| Or as another alternative one can simply run the julia source file: | ||
| - Detailed look (MITgcm) : [notebook (html)](../detailed_look/index.html) ➭ [notebook (code)](https://github.com/JuliaClimate/IndividualDisplacements.jl/blob/master/examples/basics/detailed_look.jl) | ||
|
|
||
| ``` | ||
| using IndividualDisplacements | ||
| p=dirname(pathof(IndividualDisplacements)) | ||
| include(p*"/../examples/worldwide/global_ocean_circulation.jl") | ||
| ``` | ||
|
|
||
| ## More Examples | ||
| [](https://youtu.be/W5DNqJG9jt0) | ||
|
|
||
| - Examples reproducing trajectories that had been computed earlier in Fortran ([MITgcm/pkg/flt](https://mitgcm.readthedocs.io/en/latest/outp_pkgs/outp_pkgs.html#)) are `detailed_look.jl` and `particle_cloud.jl`. | ||
| - For more examples, see: `example_CyclicArray.jl`, `example123.jl`, `helper_functions.jl`. | ||
| [](https://youtu.be/twAAE_WUs_g) |
Oops, something went wrong.